Introduction:
In today’s tutorial, we will see various techniques of cross-validation in Sklearn such as K-fold cross-validation, Stratified K-fold cross-validation, Leave one out cross-validation(LOOCV), and Repeated random train test splits. We will look into how these cross-validation techniques help better evaluation of machine learning models along with examples.
What is Cross-Validation in Machine Learning
When we create a machine learning model with supervised learning techniques it is very important to evaluate the performance and robustness of the model before moving it to production.
- How accurate the model is
- How generalized the model is
- Hold Out Approach
- Leave One Out Cross-Validation
- K-Fold Cross-Validation
- Stratified K-Fold Cross-Validation
- Repeated Random Train Test Split
1. Hold Out Approach
In the hold-out approach, the data set is split into the train and test set with random sampling. The train set is used for training the model and the test set is used to test its accuracy with unseen data. If the training and accuracy are almost the same then the model is said to have generalized well. It is common to use 80% of data for training and the remaining 20% for testing.
Advantages
- It is simple and easy to implement
- The execution time is less.
Disadvantages
- If the dataset itself is small, setting aside portions for testing would reduce the robustness of the model. This is because the training sample may not be representative of the entire dataset.
- The evaluation metrics may vary due to the randomness of the split between the train and test set.
- Although 80-20 split for train test is widely followed, there is no thumb rule for the split and hence the results can vary based on how the train test split is done.
2. Leave One Out Cross Validation (LOOCV)
In this technique, if there are n observations in the dataset, only one observation is reserved for testing, and the remaining data points are used for training. This is repeated n times till all data points have been used for testing purposes in each iteration. Finally, the average accuracy is calculated by combining the accuracies of each iteration.
Advantage
- Since every data participates both for training and testing, the overall accuracy is more reliable.
- It is very useful when the dataset is small.
Disadvantage
- LOOCV is not practical to use when the number of data observations n is huge. E.g. imagine a dataset with 500,000 records, then 500,000 model needs to be created which is not really feasible.
- There is a huge computational and time cost associated with the LOOCV approach.
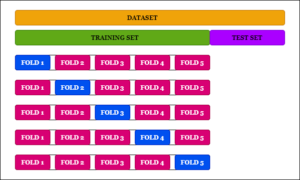
3. K-Fold Cross-Validation
In the K-Fold Cross-Validation approach, the dataset is split into K folds. Now in 1st iteration, the first fold is reserved for testing and the model is trained on the data of the remaining k-1 folds.
In the next iteration, the second fold is reserved for testing and the remaining folds are used for training. This is continued till the K-th iteration. The accuracy obtained in each iteration is used to derive the overall average accuracy for the model.
Advantages
- K-Fold cross-validation is useful when the dataset is small and splitting it is not possible to split it in train-test set (hold out approach) without losing useful data for training.
- It helps to create a robust model with low variance and low bias as it is trained on all data
Disadvantages
- The major disadvantage of K-Fold Cross Validation is that the training needs to be done K times and hence it consumes more time and resources,
- Not recommended to be used with sequential time series data.
- When the dataset is imbalanced, K-fold cross-validation may not give good results. This is because some folds may have just a few or no records for the minority class.
4. Stratified K-Fold Cross-Validation
Stratified K-fold cross-validation is useful when the data is imbalanced. While sampling data into K-folds it makes sure that the distribution of all classes in each fold is maintained. For example, if in the dataset 98% of data belongs to class B and 2% to class A, the stratified sampling will make sure each fold contains the two classes in the same ratio of 98% to 2%.
Advantage
Stratified K-fold cross-validation is recommended when the dataset is imbalanced.
5. Repeated Random Test-Train Split
Repeated random test-train split is a hybrid of traditional train-test splitting and the k-fold cross-validation method. In this technique, we create random splits of the data into the training-test set and then repeat this process multiple times, just like the cross-validation method.
Examples of Cross-Validation in Sklearn Library
About Dataset
We will be using Parkinson’s disease dataset for all examples of cross-validation in the Sklearn library. The goal is to predict whether or not a particular patient has Parkinson’s disease. We will be using the decision tree algorithm in all the examples.
The dataset has 21 attributes and 195 rows. The various fields of the Parkinson’s Disease dataset are as follows –
- MDVP:Fo(Hz) – Average vocal fundamental frequency
- MDVP:Fhi(Hz) – Maximum vocal fundamental frequency
- MDVP:Flo(Hz) – Minimum vocal fundamental frequency
- MDVP:Jitter(%),MDVP:Jitter(Abs),MDVP:RAP,MDVP:PPQ,Jitter:DDP – Several
- measures of variation in fundamental frequency
- MDVP:Shimmer,MDVP:Shimmer(dB),Shimmer:APQ3,Shimmer:APQ5,MDVP:APQ,Shimmer:DDA – Several measures of variation in amplitude
- NHR,HNR – Two measures of ratio of noise to tonal components in the voice
- status – Health status of the subject (one) – Parkinson’s, (zero) – healthy
- RPDE,D2 – Two nonlinear dynamical complexity measures
- DFA – Signal fractal scaling exponent
- spread1,spread2PPE – Three nonlinear measures of fundamental frequency variation
Also Read – Decision Tree Classifier in Python Sklearn with Example
Importing Necessary Libraries
We first load the libraries required to build our model.
import pandas as pd
import numpy as np
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
Reading CSV Data into Pandas
Next, we load the dataset in the CSV file into the pandas dataframes and check the top 5 rows.
df=pd.read_csv(r"C:\Users\Veer Kumar\Downloads\Parkinsson disease.csv")
df.head()
| name | MDVP:Fo(Hz) | MDVP:Fhi(Hz) | MDVP:Flo(Hz) | MDVP:Jitter(%) | MDVP:Jitter(Abs) | MDVP:RAP | MDVP:PPQ | Jitter:DDP | MDVP:Shimmer | … | Shimmer:DDA | NHR | HNR | status | RPDE | DFA | spread1 | spread2 | D2 | PPE | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | phon_R01_S01_1 | 119.992 | 157.302 | 74.997 | 0.00784 | 0.00007 | 0.00370 | 0.00554 | 0.01109 | 0.04374 | … | 0.06545 | 0.02211 | 21.033 | 1 | 0.414783 | 0.815285 | -4.813031 | 0.266482 | 2.301442 | 0.284654 |
| 1 | phon_R01_S01_2 | 122.400 | 148.650 | 113.819 | 0.00968 | 0.00008 | 0.00465 | 0.00696 | 0.01394 | 0.06134 | … | 0.09403 | 0.01929 | 19.085 | 1 | 0.458359 | 0.819521 | -4.075192 | 0.335590 | 2.486855 | 0.368674 |
| 2 | phon_R01_S01_3 | 116.682 | 131.111 | 111.555 | 0.01050 | 0.00009 | 0.00544 | 0.00781 | 0.01633 | 0.05233 | … | 0.08270 | 0.01309 | 20.651 | 1 | 0.429895 | 0.825288 | -4.443179 | 0.311173 | 2.342259 | 0.332634 |
| 3 | phon_R01_S01_4 | 116.676 | 137.871 | 111.366 | 0.00997 | 0.00009 | 0.00502 | 0.00698 | 0.01505 | 0.05492 | … | 0.08771 | 0.01353 | 20.644 | 1 | 0.434969 | 0.819235 | -4.117501 | 0.334147 | 2.405554 | 0.368975 |
| 4 | phon_R01_S01_5 | 116.014 | 141.781 | 110.655 | 0.01284 | 0.00011 | 0.00655 | 0.00908 | 0.01966 | 0.06425 | … | 0.10470 | 0.01767 | 19.649 | 1 | 0.417356 | 0.823484 | -3.747787 | 0.234513 | 2.332180 | 0.410335 |
5 rows × 24 columns
Data Preprocessing
The “name” column is not going to add any value in training the model and can be discarded, so we are dropping it below.
df.drop(df.columns[0], axis = 1, inplace = True)
Next, we will separate the feature and target matrix as shown below.
#Independent And dependent features
X=df.drop('status', axis=1)
y=df['status']
Hold out Approach in Sklearn
The hold-out approach can be applied by using train_test_split module of sklearn.model_selection
In the below example we have split the dataset to create the test data with a size of 30% and train data with a size of 70%. The random_state number ensures the split is deterministic in every run.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.30, random_state=4)
model = DecisionTreeClassifier()
model.fit(X_train, y_train)
result = model.score(X_test, y_test)
print(result)
0.7796610169491526
K-Fold Cross-Validation
K-Fold Cross-Validation in Sklearn can be applied by using cross_val_score module of sklearn.model_selection.
In the below example, 10 folds are used that produced 10 accuracy scores using which we calculated the mean score.
from sklearn.model_selection import cross_val_score
model=DecisionTreeClassifier()
kfold_validation=KFold(10)
results=cross_val_score(model,X,y,cv=kfold_validation)
print(results)
print(np.mean(results))
[0.7 0.8 0.8 0.8 0.8 0.78947368 0.84210526 1. 0.68421053 0.36842105] 0.758421052631579
Stratified K-fold Cross-Validation
In Sklearn stratified K-fold cross-validation can be applied by using StratifiedKFold module of sklearn.model_selection
In the below example, the dataset is divided into 5 splits or folds. It returns 5 accuracy scores using which we calculate the final mean score.
from sklearn.model_selection import StratifiedKFold
skfold=StratifiedKFold(n_splits=5)
model=DecisionTreeClassifier()
scores=cross_val_score(model,X,y,cv=skfold)
print(scores)
print(np.mean(scores))
array([0.61538462, 0.79487179, 0.71794872, 0.74358974, 0.71794872]) 0.717948717948718
Leave One Out Cross Validation(LOOCV)
In Sklearn Leave One Out Cross Validation (LOOCV) can be applied by using LeaveOneOut module of sklearn.model_selection
from sklearn.model_selection import LeaveOneOut
model=DecisionTreeClassifier()
leave_validation=LeaveOneOut()
results=cross_val_score(model,X,y,cv=leave_validation)
results
Out[22]:
array([1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 0., 0., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
0., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1.,
1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 0.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1.,
1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1.,
0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 0., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 0., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 0., 0., 1., 1., 0., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1.,
1., 1., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1., 0., 0., 1., 1., 1.,
1., 0., 1., 1., 1., 1., 1., 1., 0., 1., 1., 1., 1., 1., 1., 0., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 0.,
1., 1., 1., 1., 1., 1., 1., 1.])
print(np.mean(results))
0.8358974358974359
Repeated Random Test-Train Splits
In Sklearn repeated random test-train splits can be applied by using ShuffleSplit module of sklearn.model_selection
from sklearn.model_selection import ShuffleSplit
model=DecisionTreeClassifier()
ssplit=ShuffleSplit(n_splits=10,test_size=0.30)
results=cross_val_score(model,X,y,cv=ssplit)
print(results)
print(np.mean(results))
Out[45]:
array([0.79661017, 0.71186441, 0.79661017, 0.88135593, 0.72881356,
0.84745763, 0.83050847, 0.77966102, 0.83050847, 0.81355932])
0.8016949152542372
-
I am passionate about Analytics and I am looking for opportunities to hone my current skills to gain prominence in the field of Data Science.
View all posts


